利用molOCR截图识别文献中的结构
利用molOCR截图识别文献中的结构
手动画结构画烦了,摸会鱼。笔者是个懒狗,信奉为了偷五分钟的懒值得花上半个小时去琢磨,于是就开始搜偷懒办法。发现某商业化软件有这个功能,但一是要登陆,二是芘事多,笔者不喜欢。随后又发现了一个开源项目,拉下来之后发现还挺好用的,于是拿来玩一玩。
项目地址:molOCR
部署后端
按照说明书,该项目需要修改版的chembl_beaker作为后端,所以先装这个。由于作者提供的部署方案是docker,笔者打算直接把后端装到NAS上。
ssh连接到NAS跟着说明书完成:
1
2
3
4
5
6
7
8
9
10
11
# install docker on your linux machine
sudo snap install docker
# download
wget https://github.com/def-fun/chembl_beaker/archive/refs/heads/master.zip
unzip chembl_beaker-master.zip
# build and run
cd chembl_beaker-master
sudo docker build -f Dockerfile -t my_chembl_beaker:v1.2 .
sudo docker run -p 5000:5000 my_chembl_beaker:v1.2
docker build这一步会下载一段时间,完成后,启动容器,输出大概长这样
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
[bane@login ~/chembl_beaker-master]$ sudo docker run -p 5000:5000 my_chembl_beaker:v1.2
[07:40:10] Initializing Normalizer
CacheThrottle class can't work without cache...
Failed to load plugin beaker.plugins.throttling.Throttling because of error CacheThrottle class can't work without cache...
Caching plugin enabled but no cache backend configured, cashing will be skipped...
Bottle v0.12.18 server starting up (using GunicornServer(workers=4))...
Listening on http://0.0.0.0:5000/
Hit Ctrl-C to quit.
[2025-02-08 07:40:11 +0000] [1] [INFO] Starting gunicorn 20.0.0
[2025-02-08 07:40:11 +0000] [1] [INFO] Listening at: http://0.0.0.0:5000 (1)
[2025-02-08 07:40:11 +0000] [1] [INFO] Using worker: sync
[2025-02-08 07:40:11 +0000] [13] [INFO] Booting worker with pid: 13
[2025-02-08 07:40:12 +0000] [14] [INFO] Booting worker with pid: 14
[2025-02-08 07:40:12 +0000] [15] [INFO] Booting worker with pid: 15
[2025-02-08 07:40:12 +0000] [16] [INFO] Booting worker with pid: 16
这样后端就装好了,端口开在5000。
部署molOCR
把项目拉到本地:
1
git clone https://github.com/def-fun/molOCR.git
由于chembl_beaker服务器装在NAS上,需要先修改源码里的API接口。在js目录找到img2mol.js,找到这里:
1
2
let OCR_API_URL = 'http://' + window.location.hostname + ':5000/image2ctab'; //根据实际情况修改API的地址
// let OCR_API_URL = 'http://192.168.114.134:5000/image2ctab';
把上面一行注释掉,下面一行取消注释,IP地址改成NAS的ip:5000
1
2
// let OCR_API_URL = 'http://' + window.location.hostname + ':5000/image2ctab'; //根据实际情况修改API的地址
let OCR_API_URL = 'http://192.168.1.100:5000/image2ctab';
完成后,在项目目录启动python服务器:
1
python -m http.server
这样就启动成功了:
1
2
(base) PS D:\ChemicalPrograms\molOCR-master> python -m http.server
Serving HTTP on :: port 8000 (http://[::]:8000/) ...
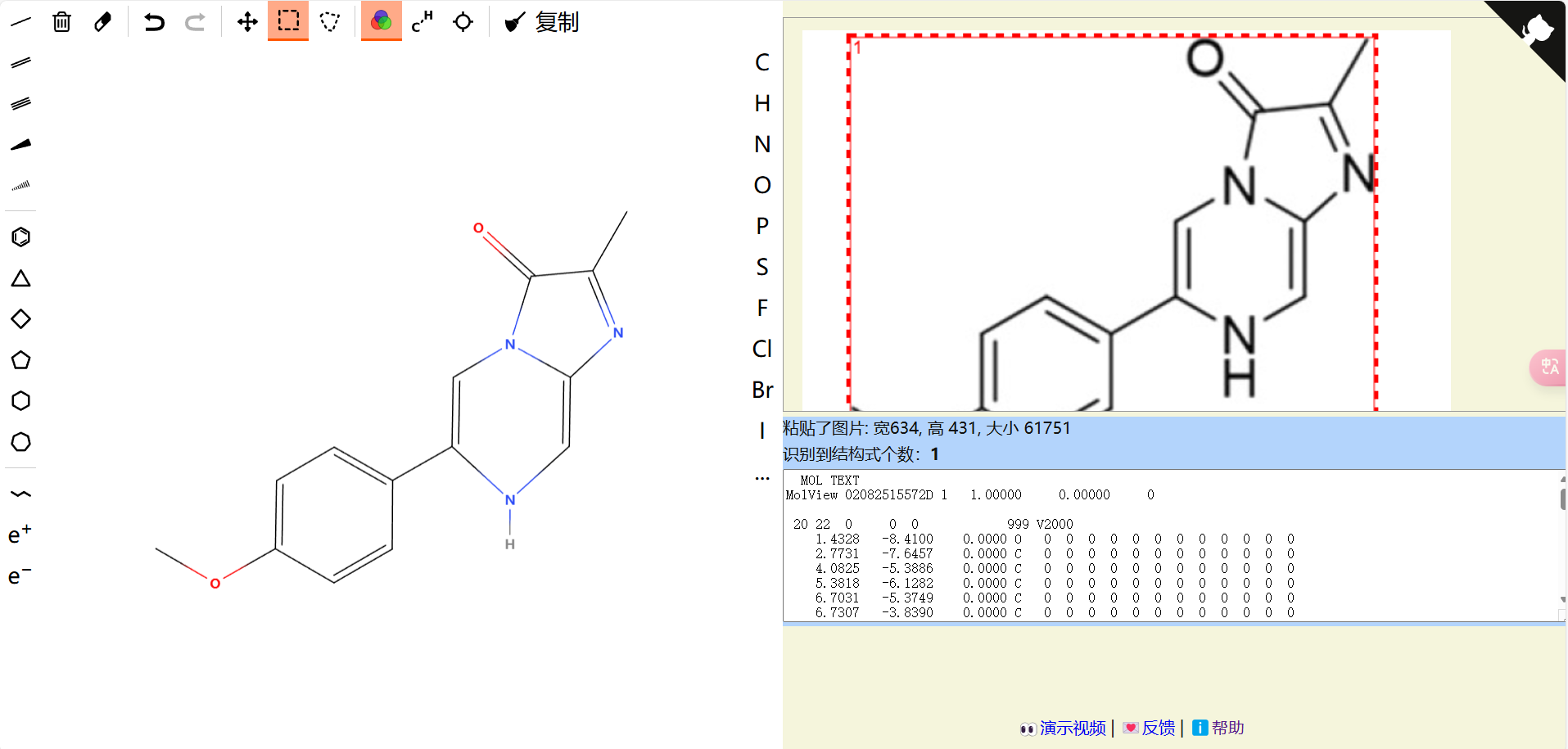
根据输出,端口是8000,打开localhost:8000,进入molOCR的页面。(笔者发现进去之后就可以关掉http服务器了,依然能运行)根据提示,可以粘贴一个截图到右侧启动识别。这笔者可太喜欢了,小键盘专门有个截图快捷键,非常方便。测试一下效果:
他这个扫把工具有可能把分子转化成同分异构体,慎用。可以点击复制粘贴到chemdraw上再clean up。
如果是打算进行量子化学计算,这个可以直接粘贴到chem3D上,或0.3版本以上的xtb-GUI上。
报错解决
failed to solve: debian:bookworm-slim
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
(obabel) [root@s02 chembl_beaker-master]# sudo docker build -f Dockerfile -t my_chembl_beaker:v1.2 .
[+] Building 3.5s (2/2) FINISHED docker:default
=> [internal] load build definition from Dockerfile 0.0s
=> => transferring dockerfile: 1.23kB 0.0s
=> ERROR [internal] load metadata for docker.io/library/debian:bookworm-slim 3.5s
------
> [internal] load metadata for docker.io/library/debian:bookworm-slim:
------
Dockerfile:1
--------------------
1 | >>> FROM debian:bookworm-slim
2 |
3 | ENV PYTHONUNBUFFERED 1
--------------------
ERROR: failed to solve: debian:bookworm-slim: failed to resolve source metadata for docker.io/library/debian:bookworm-slim: failed commit on ref "unknown-sha256:2e2509f071f9e4d6180169a868091a7b3f086c1eb87ebd42280f66471177726c": unexpected commit digest sha256:fb76fe2a60a9cc0604f31e30597ed175b83d592062772e726a9bafc2c1fdbae0, expected sha256:2e2509f071f9e4d6180169a868091a7b3f086c1eb87ebd42280f66471177726c: failed precondition
(obabel) [root@s02 chembl_beaker-master]#
Claude说这通常是由网络问题、Docker 注册表问题或 Docker 缓存数据与尝试拉取镜像之间的镜像更改引起的,推荐我:
1
docker pull debian:bookworm-slim
更新缓存后,问题解决。
This post is licensed under CC BY 4.0 by the author.